Research on soil quality is very important. Soil as a habitat for many organisms and a place where many biochemical processes occur is sensitive to natural and abiotic factors,
including agricultural activities. Soil quality assessment is based on an analysis of the physical-chemical and microbiological parameters of the soil.
In our Department, we perform the following analyses to examine the quality of the soil environment:
- soil microbial biomass carbon (MBC) and nitrogen (MBN) contents;
- dehydrogenases activity;
- acid and alkaline phosphatases activity;
- pH and EC;
- particulate organic matter (POM) content.
Soil & environmental analysis
– Soil physicochemical parameters, enzymes activity, biomass –

It is now known that only 1% of soil microorganisms can be isolated using traditional methods. For this reason, modern techniques, including molecular biology, are increasingly present in soil microbiology research. In the Department we conduct research in the scope of molecular biology and evaluation of genetic differentiation and identification of microorganisms.
One of the methods used to analysis functional diversity of soil microorganisms is the CLPP (Community Level Physiological Profiling) technique and Biolog® system, which based on it. We identification and characterization of metabolic profile of bacteria and fungi using EcoPlate, GEN II plate and FF plate.
We have the following equipment:
- Homogenizer Fast Prep®-24 (MP Biomedicals);
- NanoDrop Lite (Thermo Scientific), Spectrophotometer Evolution 60, UV-VIS spectrophotometer (Lambda 45);
- Gradient thermocycler Professional 96 Basic Gradient (Biometra), thermocycler for Real-Time PCR (AriaMix Agilent Technology);
- Sub-Cell GT Agarose Gel Electrophoresis System; gel documentation system: System Bio 1D++, Quantum ST4;
- Biolog® OmniLog® Identification System, Biolog® OmniLog® PM software, OmniLog® 2.4, MicroLog® 5.2.01, RetroSpect2 (Biolog Inc., Hayward, CA, USA).
Molecular biology, genetics
– PCR, Real Time PCR, PCR-DGGE, Biolog System –

The development and ongoing interest in methods such as metagenomics and metataxonomics enabled to broaden the knowledge of microorganisms diversity. It is estimated, however, that still 60-99% of microorganisms, called microbiological „dark matter”, remain beyond the reach of current research methods. Modern high-throughput sequencing methods used in metataxonomics make it possible to learn about microbiological diversity without the need for prior cultivation of individual species. The development of bioinformatics tools enables more precise selection of high quality reads and taxonomic identification. Assigning the correct taxonomy is the most important but also the most difficult stage in which making a mistake will result in incorrect interpretation of research results.
We offer a full bioinformatic analysis, starting with raw fastq files generated by devices such as MiSeq (llumina) and other popular sequencers.
Our pipelines include quality filtering of the reads, removing of chimeric sequences. The taxonomy can be assigned to any available an up-to-date taxonomic database including:
- RDP (Ribosome Database Project)
- SILVA
- GreenGenes (outdated)
- UNITE
- GTDB
- and others, including custom-made databases
The statistical processing includes among others:
- alpha and beta biodiversity analysis and comparisons
- graphs and plots that are ready-to-publish
- rarefaction curves
- ANOVA, PERMANOVA (…)
- Multirative analysis (PCA, PCoA, NMDS, CCA)
Metagenomics, metataxonomics
– Bioinformatics analysis, graphical & statistical processing –
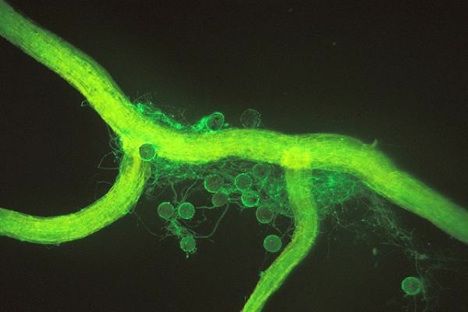
The Department also produces the bio-preparation „NITRAGINA” containing atmosferic nitogen fixing bacteria for legumes. We sell inoculates for plants such as: clover, lucerne, beans, peas, lentils, needle, vetch, soya, bean, chickpea, esparceta, acacia, lupin, peanut.
Our articles about symbiotic bacteria:
- Martyniuk S., Kozieł M., Gębala B. (2013): Response of yellow lupine to seed inoculation with Bradyrhizobium sp. (Lupinus) and with mixed inoculants of Bradyrhizobium sp. and Azotobacter chroococcum. Journal of Food Agriculture and Environment 11(2):393-396.
- Kozieł M., Gebala B., Martyniuk S. (2013): Response of Soybean to Seed Inoculation with Bradyrhizobium japonicum and with Mixed Inoculants of B. japonicum and Azotobacter chroococcum. Postępy Mikrobiologii 62(4):457-60. https://www.ncbi.nlm.nih.gov/pubmed/24730143.
- Martyniuk S., Oroń J. (2011): Use of Potato Extract Broth for Culturing Root-Nodule Bacteria. Polish Journal of Microbiology 60(4): 323-7 https://www.ncbi.nlm.nih.gov/pubmed/22390067.
- Martyniuk S., Oróń J., Martyniuk M. (2011): Diversity and numbers of root-nodule bacteria (rhizobia) in Polish soils. Acta Societatis Botanicorum Poloniae 74(1):83-86; doi: http://10.5586/asbp.2005.012.
Symbiotic bacteria
– Research on the properties, ecology and practical application of bacteria capable of atmospheric nitrogen fixation as well as symbiotic bacteria of Fabaceae –

Glomalins are soil glycoproteins produced by arbuscular mycorrhizal fungi (AMF). Glomalins are thermostable, water-insoluble glycoproteins abundantly produced by the Glomus fungi and tend to accumulate in the soil that surrounds soil aggregates and protects them from destroying. These proteins have very unique physico-chemical properties performing a fundamental role in making soil structure. It is possible that glomalins are involved in the formation of hydrophobic properties of soil, improving water and air relations in soil.
More details are available in our articles:
- Gałązka A., Gawryjołek K., Gajda A., Furtak K., Księżniak A., Jończyk K. (2018): Assessment of the glomalins content in the soil under winter wheat in different crop production systems. Plant Soil Environ., 64: 32-37. https://www.agriculturejournals.cz/publicFiles/726_2017-PSE.pdf
- Gałązka A., Gawryjołek K., Grządziel J., Księżak J. (2017): Effect of different agricultural management practices on soil biological parameters including glomalin fraction. Plant Soil Environ., 63: 300-306. https://www.agriculturejournals.cz/publicFiles/207_2017-PSE.pdf
Glomalin and glomalin-related soil proteins
– Glomalins content: total glomalin (TG), easily extractable glomalin (EEG) and glomalin-related soil proteins (GRSP) –